import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline12 Data Grouping and Aggregation
Grouping and aggregation are essential in data science because they enable meaningful analysis and insights from complex and large-scale datasets. Here are key reasons why they are important:
- Data Summarization: Grouping allows you to condense large amounts of data into concise summaries. Aggregation provides statistical summaries (e.g., mean, sum, count), making it easier to understand data trends and characteristics without needing to examine every detail.
- Insights Across Categories: Grouping by categories, like regions, demographics, or time periods, lets you analyze patterns within each group. For example, aggregating sales data by region can reveal which areas are performing better, or grouping customer data by age group can highlight consumer trends.
- Time Series Analysis: In time-series data, grouping by time intervals (e.g., day, month, quarter) allows you to observe trends over time, which can be crucial for forecasting, seasonal analysis, and trend detection.
- Reducing Data Complexity: By summarizing data at a higher level, you reduce complexity, making it easier to visualize and interpret, especially in large datasets where working with raw data could be overwhelming.
In this chapter, we are going to see grouping and aggregating using pandas. Grouping and aggregating will help to achieve data analysis easily using various functions. These methods will help us to the group and summarize our data and make complex analysis comparatively easy.
Througout this chapter, we will use gdp_lifeExpectancy.csv, let’s read the csv file to pandas dataframe first
# Load the data
gdp_lifeExp_data = pd.read_csv('./Datasets/gdp_lifeExpectancy.csv')
gdp_lifeExp_data.head()| country | continent | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | Afghanistan | Asia | 1957 | 30.332 | 9240934 | 820.853030 |
| 2 | Afghanistan | Asia | 1962 | 31.997 | 10267083 | 853.100710 |
| 3 | Afghanistan | Asia | 1967 | 34.020 | 11537966 | 836.197138 |
| 4 | Afghanistan | Asia | 1972 | 36.088 | 13079460 | 739.981106 |
12.1 Grouping by a single column
groupby() allows you to split a DataFrame based on the values in one or more columns. First, we’ll explore grouping by a single column.
12.1.1 Syntax of groupby()
DataFrame.groupby(by="column_name")– Groups data by the specified column.
Consider the life expectancy dataset. suppose we want to group by the observations by continent by passing it as an argument to the groupby() method.
#Creating a GroupBy object
grouped = gdp_lifeExp_data.groupby('continent')
#This will split the data into groups that correspond to values of the column 'continent'The groupby() method returns a GroupBy object.
#A 'GroupBy' objects is created with the `groupby()` function
type(grouped)pandas.core.groupby.generic.DataFrameGroupByThe GroupBy object grouped contains the information of the groups in which the data is distributed. Each observation has been assigned to a specific group of the column(s) used to group the data. However, note that the dataset is not physically split into different DataFrames. For example, in the above case, each observation is assigned to a particular group depending on the value of the continent for that observation. However, all the observations are still in the same DataFrame data.
12.1.2 Attributes and methods of the GroupBy object
12.1.2.1 keys
The object(s) grouping the data are called key(s). Here continent is the group key. The keys of the GroupBy object can be seen using Its keys attribute.
#Key(s) of the GroupBy object
grouped.keys'continent'12.1.2.2 ngroups
The number of groups in which the data is distributed based on the keys can be seen with the ngroups attribute.
#The number of groups based on the key(s)
grouped.ngroups5The group names are the keys of the dictionary, while the row labels are the corresponding values
#Group names
grouped.groups.keys()dict_keys(['Africa', 'Americas', 'Asia', 'Europe', 'Oceania'])12.1.2.3 groups
The groups attribute of the GroupBy object contains the group labels (or names) and the row labels of the observations in each group, as a dictionary.
#The groups (in the dictionary format)
grouped.groups{'Africa': [24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, ...], 'Americas': [48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 240, 241, 242, 243, 244, 245, 246, 247, 248, 249, 250, 251, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 286, 287, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 348, 349, 350, 351, 352, 353, 354, 355, 356, 357, 358, 359, 384, 385, 386, 387, 388, 389, 390, 391, 392, 393, 394, 395, 432, 433, 434, 435, ...], 'Asia': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 216, 217, 218, 219, 220, 221, 222, 223, 224, 225, 226, 227, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 660, 661, 662, 663, 664, 665, 666, 667, 668, 669, 670, 671, 696, 697, 698, 699, 700, 701, 702, 703, 704, 705, 706, 707, 708, 709, 710, 711, 712, 713, 714, 715, 716, 717, 718, 719, 720, 721, 722, 723, ...], 'Europe': [12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 372, 373, 374, 375, 376, 377, 378, 379, 380, 381, 382, 383, 396, 397, 398, 399, 400, 401, 402, 403, 404, 405, 406, 407, 408, 409, 410, 411, 412, 413, 414, 415, 416, 417, 418, 419, 516, 517, 518, 519, ...], 'Oceania': [60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 1092, 1093, 1094, 1095, 1096, 1097, 1098, 1099, 1100, 1101, 1102, 1103]}12.1.2.4 groups.values
The groups.values attribute of the GroupBy object contains the row labels of the observations in each group, as a dictionary.
#Group values are the row labels corresponding to a particular group
grouped.groups.values()dict_values([Index([ 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
...
1694, 1695, 1696, 1697, 1698, 1699, 1700, 1701, 1702, 1703],
dtype='int64', length=624), Index([ 48, 49, 50, 51, 52, 53, 54, 55, 56, 57,
...
1634, 1635, 1636, 1637, 1638, 1639, 1640, 1641, 1642, 1643],
dtype='int64', length=300), Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9,
...
1670, 1671, 1672, 1673, 1674, 1675, 1676, 1677, 1678, 1679],
dtype='int64', length=396), Index([ 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
...
1598, 1599, 1600, 1601, 1602, 1603, 1604, 1605, 1606, 1607],
dtype='int64', length=360), Index([ 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
1092, 1093, 1094, 1095, 1096, 1097, 1098, 1099, 1100, 1101, 1102, 1103],
dtype='int64')])12.1.2.5 size()
The size() method of the GroupBy object returns the number of observations in each group.
#Number of observations in each group
grouped.size()continent
Africa 624
Americas 300
Asia 396
Europe 360
Oceania 24
dtype: int6412.1.2.6 first()
The first non missing element of each group is returned with the first() method of the GroupBy object.
#The first element of the group can be printed using the first() method
grouped.first()| country | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|
| continent | |||||
| Africa | Algeria | 1952 | 43.077 | 9279525 | 2449.008185 |
| Americas | Argentina | 1952 | 62.485 | 17876956 | 5911.315053 |
| Asia | Afghanistan | 1952 | 28.801 | 8425333 | 779.445314 |
| Europe | Albania | 1952 | 55.230 | 1282697 | 1601.056136 |
| Oceania | Australia | 1952 | 69.120 | 8691212 | 10039.595640 |
12.1.2.7 get_group()
This method returns the observations for a particular group of the GroupBy object.
#Observations for individual groups can be obtained using the get_group() function
grouped.get_group('Asia')| country | continent | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | Afghanistan | Asia | 1957 | 30.332 | 9240934 | 820.853030 |
| 2 | Afghanistan | Asia | 1962 | 31.997 | 10267083 | 853.100710 |
| 3 | Afghanistan | Asia | 1967 | 34.020 | 11537966 | 836.197138 |
| 4 | Afghanistan | Asia | 1972 | 36.088 | 13079460 | 739.981106 |
| ... | ... | ... | ... | ... | ... | ... |
| 1675 | Yemen, Rep. | Asia | 1987 | 52.922 | 11219340 | 1971.741538 |
| 1676 | Yemen, Rep. | Asia | 1992 | 55.599 | 13367997 | 1879.496673 |
| 1677 | Yemen, Rep. | Asia | 1997 | 58.020 | 15826497 | 2117.484526 |
| 1678 | Yemen, Rep. | Asia | 2002 | 60.308 | 18701257 | 2234.820827 |
| 1679 | Yemen, Rep. | Asia | 2007 | 62.698 | 22211743 | 2280.769906 |
396 rows × 6 columns
12.2 Data aggregation within groups
12.2.1 Common Aggregation Functions
Aggregation functions are essential for summarizing and analyzing data in pandas. These functions allow you to compute summary statistics for your data, making it easier to identify trends and patterns.
Below are some of the most commonly used aggregation functions when working with grouped data in pandas:
mean()– Calculates the average value of the group.sum()– Computes the total value by summing all elements in the group.min()– Finds the minimum value in the group.max()– Finds the maximum value in the group.count()– Returns the number of occurrences or entries in the group.median()– Finds the middle value in the sorted group.std()– Calculates the standard deviation, measuring the spread or variation in the values of the group.
Each of these functions can help summarize and provide insights into different aspects of the grouped data.
Consider the life expectancy dataset, let’s find the mean life expectancy, population and GDP per capita for each country during the period of 1952 -2007.
Next, we’ll find the mean statistics for each group with the mean() method. The method will be applied on all columns of the DataFrame and all groups.
#Grouping the observations by 'country'
grouped_country = gdp_lifeExp_data.drop (['continent', 'year'], axis = 1).groupby('country')
#Finding the mean stastistic of all columns of the DataFrame and all groups
grouped_country.mean()| lifeExp | pop | gdpPercap | |
|---|---|---|---|
| country | |||
| Afghanistan | 37.478833 | 1.582372e+07 | 802.674598 |
| Albania | 68.432917 | 2.580249e+06 | 3255.366633 |
| Algeria | 59.030167 | 1.987541e+07 | 4426.025973 |
| Angola | 37.883500 | 7.309390e+06 | 3607.100529 |
| Argentina | 69.060417 | 2.860224e+07 | 8955.553783 |
| ... | ... | ... | ... |
| Vietnam | 57.479500 | 5.456857e+07 | 1017.712615 |
| West Bank and Gaza | 60.328667 | 1.848606e+06 | 3759.996781 |
| Yemen, Rep. | 46.780417 | 1.084319e+07 | 1569.274672 |
| Zambia | 45.996333 | 6.353805e+06 | 1358.199409 |
| Zimbabwe | 52.663167 | 7.641966e+06 | 635.858042 |
142 rows × 3 columns
Next, we’ll find the standard deviation statistics for each group with the std() method.
grouped_country.std()| lifeExp | pop | gdpPercap | |
|---|---|---|---|
| country | |||
| Afghanistan | 5.098646 | 7.114583e+06 | 108.202929 |
| Albania | 6.322911 | 8.285855e+05 | 1192.351513 |
| Algeria | 10.340069 | 8.613355e+06 | 1310.337656 |
| Angola | 4.005276 | 2.672281e+06 | 1165.900251 |
| Argentina | 4.186470 | 7.546609e+06 | 1862.583151 |
| ... | ... | ... | ... |
| Vietnam | 12.172331 | 2.052585e+07 | 567.482251 |
| West Bank and Gaza | 11.000069 | 1.023057e+06 | 1716.840614 |
| Yemen, Rep. | 11.019302 | 5.590408e+06 | 609.939160 |
| Zambia | 4.453246 | 3.096949e+06 | 247.494984 |
| Zimbabwe | 7.071816 | 3.376895e+06 | 133.689213 |
142 rows × 3 columns
Alternatively, you can compute other statistical metrics.
12.2.2 Multiple aggregations and Custom aggregation using agg()
12.2.2.1 Multiple aggregations
Directly applying the aggregate methods of the GroupBy object such as mean, count, etc., lets us apply only one function at a time. Also, we may wish to apply an aggregate function of our own, which is not there in the set of methods of the GroupBy object, such as the range of values of a column.
The agg() function of a GroupBy object lets us aggregate data using:
Multiple aggregation functions
Custom aggregate functions (in addition to in-built functions like mean, std, count etc.)
Consider the life expectancy dataset, Let us use the agg() method of the GroupBy object to simultaneously find the mean and standard deviation of the gdpPercap for each country.
For aggregating by multiple functions, we pass a list of strings to agg(), where the strings are the function names.
grouped_country['gdpPercap'].agg(['mean','std']).sort_values(by = 'mean',ascending = False).head()| mean | std | |
|---|---|---|
| country | ||
| Kuwait | 65332.910472 | 33882.139536 |
| Switzerland | 27074.334405 | 6886.463308 |
| Norway | 26747.306554 | 13421.947245 |
| United States | 26261.151347 | 9695.058103 |
| Canada | 22410.746340 | 8210.112789 |
12.2.2.2 Custom aggregation
In addition to the mean and standard deviation of the gdpPercap of each country, let us also include the range of gdpPercap in the table above using lambda function
grouped_country['gdpPercap'].agg(lambda x: x.max() - x.min())country
Afghanistan 342.670088
Albania 4335.973390
Algeria 3774.359280
Angola 3245.635491
Argentina 6868.064587
...
Vietnam 1836.509912
West Bank and Gaza 5595.075290
Yemen, Rep. 1499.052330
Zambia 705.723500
Zimbabwe 392.478061
Name: gdpPercap, Length: 142, dtype: float64# define a function that calculates the range
def range_func(x):
return x.max() - x.min()# apply the range function to the 'gdpPercap' column besides the mean and standard deviation
grouped_country['gdpPercap'].agg(['mean', 'std', range_func]).sort_values(by = 'range_func', ascending = False)| mean | std | range_func | |
|---|---|---|---|
| country | |||
| Kuwait | 65332.910472 | 33882.139536 | 85404.702920 |
| Singapore | 17425.382267 | 14926.147774 | 44828.041413 |
| Norway | 26747.306554 | 13421.947245 | 39261.768450 |
| Hong Kong, China | 16228.700865 | 12207.329731 | 36670.557461 |
| Ireland | 15758.606238 | 11573.311022 | 35465.716022 |
| ... | ... | ... | ... |
| Rwanda | 675.669043 | 142.229906 | 388.246772 |
| Senegal | 1533.121694 | 105.399353 | 344.572767 |
| Afghanistan | 802.674598 | 108.202929 | 342.670088 |
| Ethiopia | 509.115155 | 96.427627 | 328.659296 |
| Burundi | 471.662990 | 99.329720 | 292.403419 |
142 rows × 3 columns
For aggregating by multiple functions & changing the column names resulting from those functions, we pass a list of tuples to agg(), where each tuple is of length two, and contains the new column name & the function to be applied.
#Simultaneous renaming of columns while grouping
grouped_country['gdpPercap'].agg([('Average','mean'),('Standard Deviation','std'),('90th Percentile',lambda x:x.quantile(0.9))]).sort_values(by = '90th Percentile',ascending = False)| Average | Standard Deviation | 90th Percentile | |
|---|---|---|---|
| country | |||
| Kuwait | 65332.910472 | 33882.139536 | 109251.315590 |
| Norway | 26747.306554 | 13421.947245 | 44343.894158 |
| United States | 26261.151347 | 9695.058103 | 38764.132898 |
| Singapore | 17425.382267 | 14926.147774 | 35772.742520 |
| Switzerland | 27074.334405 | 6886.463308 | 34246.394240 |
| ... | ... | ... | ... |
| Liberia | 604.814141 | 98.988329 | 706.275527 |
| Malawi | 575.447212 | 122.999953 | 689.590541 |
| Burundi | 471.662990 | 99.329720 | 615.597260 |
| Myanmar | 439.333333 | 175.401531 | 592.300000 |
| Ethiopia | 509.115155 | 96.427627 | 577.448804 |
142 rows × 3 columns
12.2.3 Multiple aggregate functions on multiple columns
Let us find the mean and standard deviation of lifeExp and pop for each country
# find the meand and standard deviation of the 'lifeExp' and 'pop' column for each country
grouped_country[['lifeExp', 'pop']].agg(['mean', 'std']).sort_values(by = ('lifeExp', 'mean'), ascending = False)| lifeExp | pop | |||
|---|---|---|---|---|
| mean | std | mean | std | |
| country | ||||
| Iceland | 76.511417 | 3.026593 | 2.269781e+05 | 4.854168e+04 |
| Sweden | 76.177000 | 3.003990 | 8.220029e+06 | 6.365660e+05 |
| Norway | 75.843000 | 2.423994 | 4.031441e+06 | 4.107955e+05 |
| Netherlands | 75.648500 | 2.486363 | 1.378680e+07 | 2.005631e+06 |
| Switzerland | 75.565083 | 4.011572 | 6.384293e+06 | 8.582009e+05 |
| ... | ... | ... | ... | ... |
| Mozambique | 40.379500 | 4.599184 | 1.204670e+07 | 4.457509e+06 |
| Guinea-Bissau | 39.210250 | 4.937369 | 8.820084e+05 | 3.132917e+05 |
| Angola | 37.883500 | 4.005276 | 7.309390e+06 | 2.672281e+06 |
| Afghanistan | 37.478833 | 5.098646 | 1.582372e+07 | 7.114583e+06 |
| Sierra Leone | 36.769167 | 3.937828 | 3.605425e+06 | 1.270945e+06 |
142 rows × 4 columns
12.2.4 Distinct aggregate functions on multiple columns
For aggregating by multiple functions, we pass a list of strings to agg(), where the strings are the function names.
For aggregating by multiple functions & changing the column names resulting from those functions, we pass a list of tuples to agg(), where each tuple is of length two, and contains the new column name as the first object and the function to be applied as the second object of the tuple.
For aggregating by multiple functions such that a distinct set of functions is applied to each column, we pass a dictionary to agg(), where the keys are the column names on which the function is to be applied, and the values are the list of strings that are the function names, or a list of tuples if we also wish to name the aggregated columns.
# We can use a list to apply multiple aggregation functions to a single column, and a dictionary to specify different functions for multiple columns
# Use string names for the aggregation functions
grouped_country.agg({"gdpPercap": ["mean", "std"], "lifeExp": ["median", "std"], "pop": ["max", "min"]})| gdpPercap | lifeExp | pop | ||||
|---|---|---|---|---|---|---|
| mean | std | median | std | max | min | |
| country | ||||||
| Afghanistan | 802.674598 | 108.202929 | 39.1460 | 5.098646 | 31889923 | 8425333 |
| Albania | 3255.366633 | 1192.351513 | 69.6750 | 6.322911 | 3600523 | 1282697 |
| Algeria | 4426.025973 | 1310.337656 | 59.6910 | 10.340069 | 33333216 | 9279525 |
| Angola | 3607.100529 | 1165.900251 | 39.6945 | 4.005276 | 12420476 | 4232095 |
| Argentina | 8955.553783 | 1862.583151 | 69.2115 | 4.186470 | 40301927 | 17876956 |
| ... | ... | ... | ... | ... | ... | ... |
| Vietnam | 1017.712615 | 567.482251 | 57.2900 | 12.172331 | 85262356 | 26246839 |
| West Bank and Gaza | 3759.996781 | 1716.840614 | 62.5855 | 11.000069 | 4018332 | 1030585 |
| Yemen, Rep. | 1569.274672 | 609.939160 | 46.6440 | 11.019302 | 22211743 | 4963829 |
| Zambia | 1358.199409 | 247.494984 | 46.0615 | 4.453246 | 11746035 | 2672000 |
| Zimbabwe | 635.858042 | 133.689213 | 53.1765 | 7.071816 | 12311143 | 3080907 |
142 rows × 6 columns
Next, for each country, find the mean and standard deviation of the lifeExp, and the minimum and maximum values of gdpPercap.
#Specifying arguments to the function as a dictionary if distinct functions are to be applied on distinct columns
grouped_country.agg({'lifeExp':[('Average','mean'),('Standard deviation','std')],'gdpPercap':['min','max']})| lifeExp | gdpPercap | |||
|---|---|---|---|---|
| Average | Standard deviation | min | max | |
| country | ||||
| Afghanistan | 37.478833 | 5.098646 | 635.341351 | 978.011439 |
| Albania | 68.432917 | 6.322911 | 1601.056136 | 5937.029526 |
| Algeria | 59.030167 | 10.340069 | 2449.008185 | 6223.367465 |
| Angola | 37.883500 | 4.005276 | 2277.140884 | 5522.776375 |
| Argentina | 69.060417 | 4.186470 | 5911.315053 | 12779.379640 |
| ... | ... | ... | ... | ... |
| Vietnam | 57.479500 | 12.172331 | 605.066492 | 2441.576404 |
| West Bank and Gaza | 60.328667 | 11.000069 | 1515.592329 | 7110.667619 |
| Yemen, Rep. | 46.780417 | 11.019302 | 781.717576 | 2280.769906 |
| Zambia | 45.996333 | 4.453246 | 1071.353818 | 1777.077318 |
| Zimbabwe | 52.663167 | 7.071816 | 406.884115 | 799.362176 |
142 rows × 4 columns
12.3 Grouping by Multiple Columns
Above, we demonstrated grouping by a single column, which is useful for summarizing data based on one categorical variable. However, in many cases, we need to group by multiple columns. Grouping by multiple columns allows us to create more detailed summaries by accounting for multiple categorical variables. This approach enables us to analyze data at a finer granularity, revealing insights that might be missed with single-column grouping alone.
12.3.1 Basic Syntax for Grouping by Multiple Columns
Use groupby() with a list of column names to group data by multiple columns.
DataFrame.groupby(by=["col1", "col2"])
Consider the life expectancy dataset, we can group by both country and continent to analyze gdpPercap, lifeExp, and pop trends for each country within each continent, providing a more comprehensive view of the data.
#Grouping by multiple columns
grouped_continent_contry = gdp_lifeExp_data.groupby(['continent', 'country'])[ "lifeExp"].agg(['mean', 'std', 'max', 'min']).sort_values(by = 'mean', ascending = False)grouped_continent_contry| mean | std | max | min | ||
|---|---|---|---|---|---|
| continent | country | ||||
| Europe | Iceland | 76.511417 | 3.026593 | 81.757 | 72.490 |
| Sweden | 76.177000 | 3.003990 | 80.884 | 71.860 | |
| Norway | 75.843000 | 2.423994 | 80.196 | 72.670 | |
| Netherlands | 75.648500 | 2.486363 | 79.762 | 72.130 | |
| Switzerland | 75.565083 | 4.011572 | 81.701 | 69.620 | |
| ... | ... | ... | ... | ... | ... |
| Africa | Mozambique | 40.379500 | 4.599184 | 46.344 | 31.286 |
| Guinea-Bissau | 39.210250 | 4.937369 | 46.388 | 32.500 | |
| Angola | 37.883500 | 4.005276 | 42.731 | 30.015 | |
| Asia | Afghanistan | 37.478833 | 5.098646 | 43.828 | 28.801 |
| Africa | Sierra Leone | 36.769167 | 3.937828 | 42.568 | 30.331 |
142 rows × 4 columns
12.3.2 Understanding Hierarchical (Multi-Level) Indexing
- Grouping by multiple columns creates a hierarchical index (also called a multi-level index).
- This index allows each level (e.g., continent, country) to act as an independent category that can be accessed individually.
In the above output, continent and country form a two-level hierarchical index, allowing us to drill down from continent-level to country-level summaries.
grouped_continent_contry.index.nlevels2# get the first level of the index
grouped_continent_contry.index.levels[0]Index(['Africa', 'Americas', 'Asia', 'Europe', 'Oceania'], dtype='object', name='continent')# get the second level of the index
grouped_continent_contry.index.levels[1]Index(['Afghanistan', 'Albania', 'Algeria', 'Angola', 'Argentina', 'Australia',
'Austria', 'Bahrain', 'Bangladesh', 'Belgium',
...
'Uganda', 'United Kingdom', 'United States', 'Uruguay', 'Venezuela',
'Vietnam', 'West Bank and Gaza', 'Yemen, Rep.', 'Zambia', 'Zimbabwe'],
dtype='object', name='country', length=142)12.3.3 Subsetting Data in a Hierarchical Index
grouped_continent_country is still a DataFrame with hierarchical indexing. You can use .loc[] for subsetting, just as you would with a single-level index.
# get the observations for the 'Americas' continent
grouped_continent_contry.loc['Americas'].head()| mean | std | max | min | |
|---|---|---|---|---|
| country | ||||
| Canada | 74.902750 | 3.952871 | 80.653 | 68.750 |
| United States | 73.478500 | 3.343781 | 78.242 | 68.440 |
| Puerto Rico | 72.739333 | 3.984267 | 78.746 | 64.280 |
| Cuba | 71.045083 | 6.022798 | 78.273 | 59.421 |
| Uruguay | 70.781583 | 3.342937 | 76.384 | 66.071 |
# get the mean life expectancy for the 'Americas' continent
grouped_continent_contry.loc['Americas']['mean'].head()country
Canada 74.902750
United States 73.478500
Puerto Rico 72.739333
Cuba 71.045083
Uruguay 70.781583
Name: mean, dtype: float64# another way to get the mean life expectancy for the 'Americas' continent
grouped_continent_contry.loc['Americas', 'mean'].head()country
Canada 74.902750
United States 73.478500
Puerto Rico 72.739333
Cuba 71.045083
Uruguay 70.781583
Name: mean, dtype: float64You can use a tuple to access data for specific levels in a multi-level index.
# get the observations for the 'United States' country
grouped_continent_contry.loc[( 'Americas', 'United States')]mean 73.478500
std 3.343781
max 78.242000
min 68.440000
Name: (Americas, United States), dtype: float64grouped_continent_contry.loc[( 'Americas', 'United States'), ['mean', 'std']]mean 73.478500
std 3.343781
Name: (Americas, United States), dtype: float64gdp_lifeExp_data.columnsIndex(['country', 'continent', 'year', 'lifeExp', 'pop', 'gdpPercap'], dtype='object')Finally, you can use reset_index() to convert the hierarchical index into a regular index, allowing you to apply the standard subsetting and filtering methods covered in previous chapters
grouped_continent_contry.reset_index().head()| continent | country | mean | std | max | min | |
|---|---|---|---|---|---|---|
| 0 | Europe | Iceland | 76.511417 | 3.026593 | 81.757 | 72.49 |
| 1 | Europe | Sweden | 76.177000 | 3.003990 | 80.884 | 71.86 |
| 2 | Europe | Norway | 75.843000 | 2.423994 | 80.196 | 72.67 |
| 3 | Europe | Netherlands | 75.648500 | 2.486363 | 79.762 | 72.13 |
| 4 | Europe | Switzerland | 75.565083 | 4.011572 | 81.701 | 69.62 |
12.3.4 Grouping by multiple columns and aggregating multiple variables
#Grouping by multiple columns
grouped_continent_contry_multi = gdp_lifeExp_data.groupby(['continent', 'country','year'])[ ['lifeExp', 'pop', 'gdpPercap']].agg(['mean', 'max', 'min'])
grouped_continent_contry_multi| lifeExp | pop | gdpPercap | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | max | min | mean | max | min | mean | max | min | |||
| continent | country | year | |||||||||
| Africa | Algeria | 1952 | 43.077 | 43.077 | 43.077 | 9279525.0 | 9279525 | 9279525 | 2449.008185 | 2449.008185 | 2449.008185 |
| 1957 | 45.685 | 45.685 | 45.685 | 10270856.0 | 10270856 | 10270856 | 3013.976023 | 3013.976023 | 3013.976023 | ||
| 1962 | 48.303 | 48.303 | 48.303 | 11000948.0 | 11000948 | 11000948 | 2550.816880 | 2550.816880 | 2550.816880 | ||
| 1967 | 51.407 | 51.407 | 51.407 | 12760499.0 | 12760499 | 12760499 | 3246.991771 | 3246.991771 | 3246.991771 | ||
| 1972 | 54.518 | 54.518 | 54.518 | 14760787.0 | 14760787 | 14760787 | 4182.663766 | 4182.663766 | 4182.663766 | ||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Oceania | New Zealand | 1987 | 74.320 | 74.320 | 74.320 | 3317166.0 | 3317166 | 3317166 | 19007.191290 | 19007.191290 | 19007.191290 |
| 1992 | 76.330 | 76.330 | 76.330 | 3437674.0 | 3437674 | 3437674 | 18363.324940 | 18363.324940 | 18363.324940 | ||
| 1997 | 77.550 | 77.550 | 77.550 | 3676187.0 | 3676187 | 3676187 | 21050.413770 | 21050.413770 | 21050.413770 | ||
| 2002 | 79.110 | 79.110 | 79.110 | 3908037.0 | 3908037 | 3908037 | 23189.801350 | 23189.801350 | 23189.801350 | ||
| 2007 | 80.204 | 80.204 | 80.204 | 4115771.0 | 4115771 | 4115771 | 25185.009110 | 25185.009110 | 25185.009110 | ||
1704 rows × 9 columns
Breaking Down Grouping and Aggregation
Grouping by Multiple Columns:
In this example, we are grouping the data by three columns:continent,country, andyear. This creates groups based on unique combinations of these columns.Aggregating Multiple Variables:
We apply multiple aggregation functions (mean,std,max, andmin) to multiple variables (lifeExp,pop, andgdpPercap).
This type of operation is commonly referred to as “multi-column grouping with multiple aggregations” in pandas. It’s a powerful approach because it allows us to obtain a detailed statistical summary for each combination of grouping columns across several variables.
# its columns are also two levels deep
grouped_continent_contry_multi.columns.nlevels2# pass a tuple to the loc() method to access the values of the multi-level columns with a multi-level index
grouped_continent_contry_multi.loc[('Americas','United States'), ('lifeExp', 'mean')]year
1952 68.440
1957 69.490
1962 70.210
1967 70.760
1972 71.340
1977 73.380
1982 74.650
1987 75.020
1992 76.090
1997 76.810
2002 77.310
2007 78.242
Name: (lifeExp, mean), dtype: float6412.4 Advanced Operations within groups: apply(), transform(), and filter()
12.4.1 Using apply() on groups
The apply() function applies a custom function to each group, allowing for flexible operations. The function can return either a scalar, Series, or DataFrame.
Example: Consider the life expectancy dataset, find the top 3 life expectancy values for each continent
We’ll first define a function that sorts a dataset by decreasing life expectancy and returns the top 3 rows. Then, we’ll apply this function on each group using the apply() method of the GroupBy object.
# Define a function to get the top 3 rows based on life expectancy for each group
def top_3_life_expectancy(group):
return group.nlargest(3, 'lifeExp')#Defining the groups in the data
grouped_gdpcapital_data = gdp_lifeExp_data.groupby('continent')Now we’ll use the apply() method to apply the top_3_life_expectancy() function on each group of the object grouped_gdpcapital_data.
# Apply the function to each continent group
top_life_expectancy = gdp_lifeExp_data.groupby('continent')[['continent', 'country', 'year', 'lifeExp', 'gdpPercap']].apply(top_3_life_expectancy).reset_index(drop=True)
# Display the result
top_life_expectancy.head()| continent | country | year | lifeExp | gdpPercap | |
|---|---|---|---|---|---|
| 0 | Africa | Reunion | 2007 | 76.442 | 7670.122558 |
| 1 | Africa | Reunion | 2002 | 75.744 | 6316.165200 |
| 2 | Africa | Reunion | 1997 | 74.772 | 6071.941411 |
| 3 | Americas | Canada | 2007 | 80.653 | 36319.235010 |
| 4 | Americas | Canada | 2002 | 79.770 | 33328.965070 |
The top_3_life_expectancy() function is applied to each group, and the results are concatenated internally with the concat() function. The output therefore has a hierarchical index whose outer level indices are the group keys.
We can also use a lambda function instead of separately defining the function top_3_life_expectancy():
# Use a lambda function to get the top 3 life expectancy values for each continent
top_life_expectancy = (
gdp_lifeExp_data
.groupby('continent')[['continent', 'country', 'year', 'lifeExp', 'gdpPercap']] # Avoid adding group labels in the index
.apply(lambda x: x.nlargest(3, 'lifeExp'))
.reset_index(drop=True)
)
# Display the result
top_life_expectancy.head()| continent | country | year | lifeExp | gdpPercap | |
|---|---|---|---|---|---|
| 0 | Africa | Reunion | 2007 | 76.442 | 7670.122558 |
| 1 | Africa | Reunion | 2002 | 75.744 | 6316.165200 |
| 2 | Africa | Reunion | 1997 | 74.772 | 6071.941411 |
| 3 | Americas | Canada | 2007 | 80.653 | 36319.235010 |
| 4 | Americas | Canada | 2002 | 79.770 | 33328.965070 |
12.4.2 Using transform() on Groups
The transform() function applies a function to each group and returns a Series aligned with the original DataFrame’s index. This makes it suitable for adding or modifying columns based on group-level calculations.
Recall that in the data cleaning and preparation chapter, we imputed missing values based on correlated variables in the dataset.
In the example provided, some countries had missing values for GDP per capita. To handle this, we imputed the missing GDP per capita for each country using the average GDP per capita of its corresponding continent.
Now, we’ll explore an alternative approach using groupby() and transform() to perform this imputation.
Let us read the datasets and the function that makes a visualization to compare the imputed values with the actual values.
#Importing data with missing values
gdp_missing_data = pd.read_csv('./Datasets/GDP_missing_data.csv')
#Importing data with all values
gdp_complete_data = pd.read_csv('./Datasets/GDP_complete_data.csv')#Index of rows with missing values for GDP per capita
null_ind_gdpPC = gdp_missing_data.index[gdp_missing_data.gdpPerCapita.isnull()]
#Defining a function to plot the imputed values vs actual values
def plot_actual_vs_predicted():
fig, ax = plt.subplots(figsize=(8, 6))
plt.rc('xtick', labelsize=15)
plt.rc('ytick', labelsize=15)
x = gdp_complete_data.loc[null_ind_gdpPC,'gdpPerCapita']
y = gdp_imputed_data.loc[null_ind_gdpPC,'gdpPerCapita']
plt.scatter(x,y)
z=np.polyfit(x,y,1)
p=np.poly1d(z)
plt.plot(x,x,color='orange')
plt.xlabel('Actual GDP per capita',fontsize=20)
plt.ylabel('Imputed GDP per capita',fontsize=20)
ax.xaxis.set_major_formatter('${x:,.0f}')
ax.yaxis.set_major_formatter('${x:,.0f}')
rmse = np.sqrt(((x-y).pow(2)).mean())
print("RMSE=",rmse)Approach 1: Using the approach we used in the previous chapter
#Finding the mean GDP per capita of the continent
avg_gdpPerCapita = gdp_missing_data['gdpPerCapita'].groupby(gdp_missing_data['continent']).mean()
#Creating a copy of missing data to impute missing values
gdp_imputed_data = gdp_missing_data.copy()
#Replacing missing GDP per capita with the mean GDP per capita for the corresponding continent
for cont in avg_gdpPerCapita.index:
gdp_imputed_data.loc[(gdp_imputed_data.continent==cont) & (gdp_imputed_data.gdpPerCapita.isnull()),
'gdpPerCapita']=avg_gdpPerCapita[cont]
plot_actual_vs_predicted()RMSE= 25473.20645170116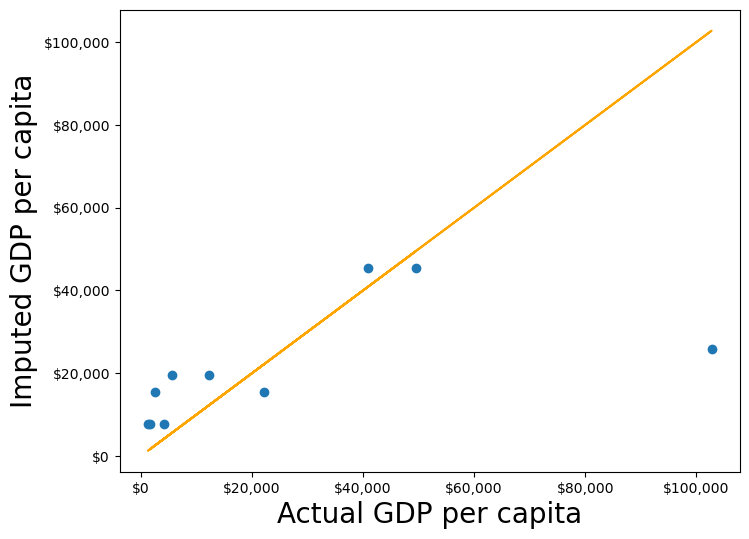
Approach 2: Using the groupby() and transform() methods.
The transform() function is a powerful tool for filling missing values in grouped data. It allows us to apply a function across each group and align the result back to the original DataFrame, making it perfect for filling missing values based on group statistics.
In this example, we use transform() to impute missing values in the gdpPerCapita column by filling them with the mean gdpPerCapita of each continent:
#Creating a copy of missing data to impute missing values
gdp_imputed_data = gdp_missing_data.copy()
#Grouping data by continent
grouped = gdp_missing_data.groupby('continent')
#Imputing missing values with the mean GDP per capita of the continent
gdp_imputed_data['gdpPerCapita'] = grouped['gdpPerCapita'].transform(lambda x: x.fillna(x.mean()))
plot_actual_vs_predicted()RMSE= 25473.20645170116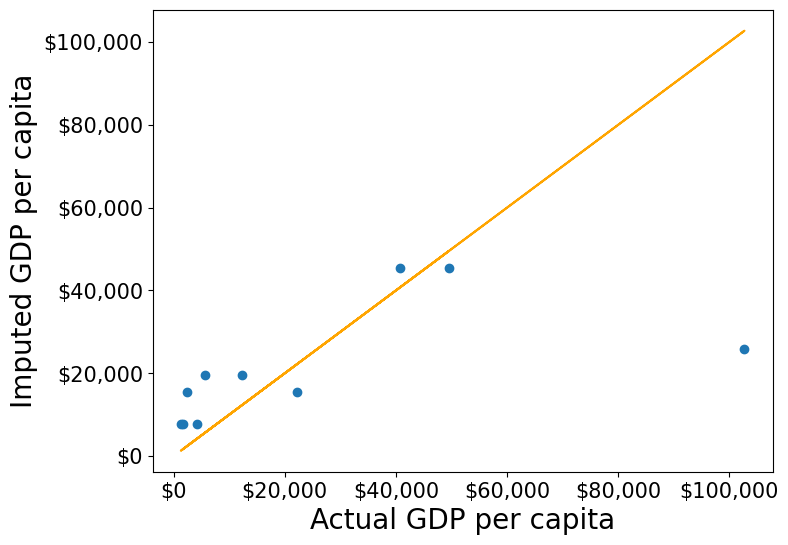
Using the transform() function, missing values in gdpPerCapita for each group are filled with the group’s mean gdpPerCapita. This approach is not only more convenient to write but also faster compared to using for loops. While a for loop imputes missing values one group at a time, transform() performs built-in operations (like mean, sum, etc.) in a way that is optimized internally, making it more efficient.
Let’s use apply() instead of transform() with groupby()
#Creating a copy of missing data to impute missing values
gdp_imputed_data = gdp_missing_data.copy()
#Grouping data by continent
grouped = gdp_missing_data.groupby('continent')
#Applying the lambda function on the 'gdpPerCapita' column of the groups
gdp_imputed_data['gdpPerCapita'] = grouped['gdpPerCapita'].apply(lambda x: x.fillna(x.mean()))
plot_actual_vs_predicted()TypeError: incompatible index of inserted column with frame indexWhy we ran into this error? and apply() doesn’t work?
Here’s a deeper look at why apply() doesn’t work as expected here:
12.4.2.1 Behavior of groupby().apply() vs. groupby().transform()
groupby().apply(): This method applies a function to each group and returns the result with a hierarchical (multi-level) index by default. This hierarchical index can make it difficult to align the result back to a single column in the original DataFrame.groupby().transform(): In contrast,transform()is specifically designed to apply a function to each group and return a Series that is aligned with the original DataFrame’s index. This alignment makes it directly compatible for assignment to a new or existing column in the original DataFrame.
12.4.2.2 Why transform() Works for Imputation
When using transform() to fill missing values, it applies the function (e.g., fillna(x.mean())) based on each group’s statistics, such as the mean, while keeping the result aligned with the original DataFrame’s index. This allows for smooth assignment to a column in the DataFrame without any index mismatch issues.
Additionally, transform() applies the function to each element in a group independently and returns a result that has the same shape as the original data, making it ideal for adding or modifying columns.
12.4.3 Using filter() on Groups
The filter() function filters entire groups based on a condition. It evaluates each group and keeps only those that meet the specified criteria.
Example: Keep only the countries where the mean life expectancy is greater than 70
# keep only the continent where the mean life expectancy is greater than 74
gdp_lifeExp_data.groupby('continent').filter(lambda x: x['lifeExp'].mean() > 74)['continent'].unique()array(['Oceania'], dtype=object)# keep only the country where the mean life expectancy is greater than 74
gdp_lifeExp_data.groupby('country').filter(lambda x: x['lifeExp'].mean() > 74)['country'].unique()array(['Australia', 'Canada', 'Denmark', 'France', 'Iceland', 'Italy',
'Japan', 'Netherlands', 'Norway', 'Spain', 'Sweden', 'Switzerland'],
dtype=object)Using .nunique() get the number of countries that satisfy this condition
gdp_lifeExp_data.groupby('country').filter(lambda x: x['lifeExp'].mean() > 74)['country'].nunique()1212.5 Sampling data by group
If a dataset contains a large number of observations, operating on it can be computationally expensive. Instead, working on a sample of entire observations is a more efficient alterative. The groupby() method combined with apply() can be used for stratified random sampling from a large dataset.
Before taking the random sample, let us find the number of countries in each continent.
gdp_lifeExp_data.continent.value_counts()continent
Africa 624
Asia 396
Europe 360
Americas 300
Oceania 24
Name: count, dtype: int64Let us take a random sample of 650 observations from the entire dataset.
sample_lifeExp_data = gdp_lifeExp_data.sample(650)Now, let us see the number of countries of each continent in our sample.
sample_lifeExp_data.continent.value_counts()continent
Africa 241
Asia 149
Europe 142
Americas 109
Oceania 9
Name: count, dtype: int64Some of the continent have a very low representation in the data. To rectify this, we can take a random sample of 130 observations from each of the 5 continents. In other words, we can take a random sample from each of the continent-based groups.
evenly_sampled_lifeExp_data = gdp_lifeExp_data.groupby('continent').apply(lambda x:x.sample(130, replace=True), include_groups=False)
group_sizes = evenly_sampled_lifeExp_data.groupby(level=0).size()
print(group_sizes)continent
Africa 130
Americas 130
Asia 130
Europe 130
Oceania 130
dtype: int64The above stratified random sample equally represents all the continent.
12.6 corr(): Correlation by group
The corr() method of the GroupBy object returns the correlation between all pairs of columns within each group.
Example: Find the correlation between lifeExp and gdpPercap for each continent-country level combination.
gdp_lifeExp_data.groupby(['continent','country']).apply(lambda x:x['lifeExp'].corr(x['gdpPercap']), include_groups=False)continent country
Africa Algeria 0.904471
Angola -0.301079
Benin 0.843949
Botswana 0.005597
Burkina Faso 0.881677
...
Europe Switzerland 0.980715
Turkey 0.954455
United Kingdom 0.989893
Oceania Australia 0.986446
New Zealand 0.974493
Length: 142, dtype: float64Life expectancy is closely associated with GDP per capita across most continent-country combinations.
12.7 pivot_table()
The pivot_table() function in pandas is a powerful tool for performing groupwise aggregation in a structured format, similar to Excel’s pivot tables. It allows you to create a summary of data by grouping and aggregating it based on specified columns. Here’s an overview of how pivot_table() works for groupwise aggregation:
Note that pivot_table() is the same as pivot() except that pivot_table() aggregates the data as well in addition to re-arranging it.
Example: Consider the life expectancy dataset, calculate the average life expectancy for each country and year combination
pd.pivot_table(data = gdp_lifeExp_data, values = 'lifeExp',index = 'country', columns ='year',aggfunc = 'mean',margins = True)| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 | All |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| country | |||||||||||||
| Afghanistan | 28.80100 | 30.332000 | 31.997000 | 34.02000 | 36.088000 | 38.438000 | 39.854000 | 40.822000 | 41.674000 | 41.763000 | 42.129000 | 43.828000 | 37.478833 |
| Albania | 55.23000 | 59.280000 | 64.820000 | 66.22000 | 67.690000 | 68.930000 | 70.420000 | 72.000000 | 71.581000 | 72.950000 | 75.651000 | 76.423000 | 68.432917 |
| Algeria | 43.07700 | 45.685000 | 48.303000 | 51.40700 | 54.518000 | 58.014000 | 61.368000 | 65.799000 | 67.744000 | 69.152000 | 70.994000 | 72.301000 | 59.030167 |
| Angola | 30.01500 | 31.999000 | 34.000000 | 35.98500 | 37.928000 | 39.483000 | 39.942000 | 39.906000 | 40.647000 | 40.963000 | 41.003000 | 42.731000 | 37.883500 |
| Argentina | 62.48500 | 64.399000 | 65.142000 | 65.63400 | 67.065000 | 68.481000 | 69.942000 | 70.774000 | 71.868000 | 73.275000 | 74.340000 | 75.320000 | 69.060417 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| West Bank and Gaza | 43.16000 | 45.671000 | 48.127000 | 51.63100 | 56.532000 | 60.765000 | 64.406000 | 67.046000 | 69.718000 | 71.096000 | 72.370000 | 73.422000 | 60.328667 |
| Yemen, Rep. | 32.54800 | 33.970000 | 35.180000 | 36.98400 | 39.848000 | 44.175000 | 49.113000 | 52.922000 | 55.599000 | 58.020000 | 60.308000 | 62.698000 | 46.780417 |
| Zambia | 42.03800 | 44.077000 | 46.023000 | 47.76800 | 50.107000 | 51.386000 | 51.821000 | 50.821000 | 46.100000 | 40.238000 | 39.193000 | 42.384000 | 45.996333 |
| Zimbabwe | 48.45100 | 50.469000 | 52.358000 | 53.99500 | 55.635000 | 57.674000 | 60.363000 | 62.351000 | 60.377000 | 46.809000 | 39.989000 | 43.487000 | 52.663167 |
| All | 49.05762 | 51.507401 | 53.609249 | 55.67829 | 57.647386 | 59.570157 | 61.533197 | 63.212613 | 64.160338 | 65.014676 | 65.694923 | 67.007423 | 59.474439 |
143 rows × 13 columns
Explanation
- values: Specifies the column to aggregate (e.g.,
lifeExpin our example). - index: Groups by the rows based on the
countrycolumn. - columns: Groups by the columns based on the
yearcolumn. - aggfunc: Uses
meanto calculate the average life expectancy.
Common Aggregation Functions in pivot_table()
You can use various aggregation functions within pivot_table() to summarize data:
mean– Calculates the average of values within each group.sum– Computes the total of values within each group.count– Counts the number of non-null entries within each group.minandmax– Finds the minimum and maximum values within each group.
We can also use custom GroupBy aggregate functions with pivot_table().
Example: Find the \(90^{th}\) percentile of life expectancy for each country and year combination
pd.pivot_table(data = gdp_lifeExp_data, values = 'lifeExp',index = 'country', columns ='year',aggfunc = lambda x:np.percentile(x,90))| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| country | ||||||||||||
| Afghanistan | 28.801 | 30.332 | 31.997 | 34.020 | 36.088 | 38.438 | 39.854 | 40.822 | 41.674 | 41.763 | 42.129 | 43.828 |
| Albania | 55.230 | 59.280 | 64.820 | 66.220 | 67.690 | 68.930 | 70.420 | 72.000 | 71.581 | 72.950 | 75.651 | 76.423 |
| Algeria | 43.077 | 45.685 | 48.303 | 51.407 | 54.518 | 58.014 | 61.368 | 65.799 | 67.744 | 69.152 | 70.994 | 72.301 |
| Angola | 30.015 | 31.999 | 34.000 | 35.985 | 37.928 | 39.483 | 39.942 | 39.906 | 40.647 | 40.963 | 41.003 | 42.731 |
| Argentina | 62.485 | 64.399 | 65.142 | 65.634 | 67.065 | 68.481 | 69.942 | 70.774 | 71.868 | 73.275 | 74.340 | 75.320 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Vietnam | 40.412 | 42.887 | 45.363 | 47.838 | 50.254 | 55.764 | 58.816 | 62.820 | 67.662 | 70.672 | 73.017 | 74.249 |
| West Bank and Gaza | 43.160 | 45.671 | 48.127 | 51.631 | 56.532 | 60.765 | 64.406 | 67.046 | 69.718 | 71.096 | 72.370 | 73.422 |
| Yemen, Rep. | 32.548 | 33.970 | 35.180 | 36.984 | 39.848 | 44.175 | 49.113 | 52.922 | 55.599 | 58.020 | 60.308 | 62.698 |
| Zambia | 42.038 | 44.077 | 46.023 | 47.768 | 50.107 | 51.386 | 51.821 | 50.821 | 46.100 | 40.238 | 39.193 | 42.384 |
| Zimbabwe | 48.451 | 50.469 | 52.358 | 53.995 | 55.635 | 57.674 | 60.363 | 62.351 | 60.377 | 46.809 | 39.989 | 43.487 |
142 rows × 12 columns
12.8 crosstab()
12.8.1 Basic Usage of crosstab()
The crosstab() method is a special case of a pivot table for computing group frequncies (or size of each group).
# create a basic crosstab to see counts of countries in each continent
pd.crosstab(gdp_lifeExp_data['continent'], columns='count')| col_0 | count |
|---|---|
| continent | |
| Africa | 624 |
| Americas | 300 |
| Asia | 396 |
| Europe | 360 |
| Oceania | 24 |
Use the margins=True argument to add totals (row and column margins) to the table. The All row and column provide totals for each age group and gender category.
# create a crosstab to see counts of countries in each continent and year
pd.crosstab(gdp_lifeExp_data['continent'], gdp_lifeExp_data['year'], margins=True)| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 | All |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| continent | |||||||||||||
| Africa | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 52 | 624 |
| Americas | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 25 | 300 |
| Asia | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 396 |
| Europe | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 360 |
| Oceania | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 24 |
| All | 142 | 142 | 142 | 142 | 142 | 142 | 142 | 142 | 142 | 142 | 142 | 142 | 1704 |
This table shows the count of each year in each continent group, helping us understand the year distribution across different continent groups. We may often use it to check if the data is representative of all groups that are of interest to us.
12.8.2 Using crosstab() with Aggregation Functions
You can specify a values column and an aggregation function (aggfunc) to summarize numerical data for each combination of categorical variables.
Example: find the mean life expectancy for each continent and year
# find the mean life expectancy for each country in each continent and year
pd.crosstab(gdp_lifeExp_data['continent'], gdp_lifeExp_data['year'], values=gdp_lifeExp_data['lifeExp'], aggfunc='mean')| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| continent | ||||||||||||
| Africa | 39.135500 | 41.266346 | 43.319442 | 45.334538 | 47.450942 | 49.580423 | 51.592865 | 53.344788 | 53.629577 | 53.598269 | 53.325231 | 54.806038 |
| Americas | 53.279840 | 55.960280 | 58.398760 | 60.410920 | 62.394920 | 64.391560 | 66.228840 | 68.090720 | 69.568360 | 71.150480 | 72.422040 | 73.608120 |
| Asia | 46.314394 | 49.318544 | 51.563223 | 54.663640 | 57.319269 | 59.610556 | 62.617939 | 64.851182 | 66.537212 | 68.020515 | 69.233879 | 70.728485 |
| Europe | 64.408500 | 66.703067 | 68.539233 | 69.737600 | 70.775033 | 71.937767 | 72.806400 | 73.642167 | 74.440100 | 75.505167 | 76.700600 | 77.648600 |
| Oceania | 69.255000 | 70.295000 | 71.085000 | 71.310000 | 71.910000 | 72.855000 | 74.290000 | 75.320000 | 76.945000 | 78.190000 | 79.740000 | 80.719500 |
12.9 Independent Study
12.9.1 Practice exercise 1
Read the table consisting of GDP per capita of countries from the webpage: https://en.wikipedia.org/wiki/List_of_countries_by_GDP_(nominal)_per_capita .
To only read the relevant table, read the tables that contain the word ‘Country’.
Estimate the GDP per capita of each country as the average of the estimates of the three agencies - IMF, United Nations and World Bank.
We need to do a bit of data cleaning before we could directly use the groupby() function. Follow the steps below:
- Drop the “Year” column
- Drop the level 1 column name (innermost level column)
- Apply the following function on all the columns of the “Estimate” to convert them to numeric:
f = lambda x:pd.to_numeric(x,errors = 'coerce') - Set the Country/Territory column as the index
- Convert all other columns into numeric and coerce errors using ‘errors=’coerce’`
- Drop rows with NaN values
- Find the average GDP per capital across the three agencies
dfs = pd.read_html('https://en.wikipedia.org/wiki/List_of_countries_by_GDP_(nominal)_per_capita', match = 'Country')
gdp_data = dfs[0]
gdp_data.head()| Country/Territory | IMF[4][5] | World Bank[6] | United Nations[7] | ||||
|---|---|---|---|---|---|---|---|
| Country/Territory | Estimate | Year | Estimate | Year | Estimate | Year | |
| 0 | Monaco | — | — | 240862 | 2022 | 240535 | 2022 |
| 1 | Liechtenstein | — | — | 187267 | 2022 | 197268 | 2022 |
| 2 | Luxembourg | 135321 | 2024 | 128259 | 2023 | 125897 | 2022 |
| 3 | Bermuda | — | — | 123091 | 2022 | 117568 | 2022 |
| 4 | Switzerland | 106098 | 2024 | 99995 | 2023 | 93636 | 2022 |
# Flatten the MultiIndex to access column names
gdp_data.columns = [' '.join(col).strip() if isinstance(col, tuple) else col for col in gdp_data.columns]
# Drop all columns containing "Year"
gdp_data = gdp_data.loc[:, ~gdp_data.columns.str.contains('Year', case=False)]
gdp_data = gdp_data.rename(columns={"Country/Territory Country/Territory": "Country/Territory"})
gdp_data.head()| Country/Territory | IMF | World Bank | United Nations | |
|---|---|---|---|---|
| 0 | Monaco | — | 240862 | 240535 |
| 1 | Liechtenstein | — | 187267 | 197268 |
| 2 | Luxembourg | 135321 | 128259 | 125897 |
| 3 | Bermuda | — | 123091 | 117568 |
| 4 | Switzerland | 106098 | 99995 | 93636 |
import re
column_name_cleaner = lambda x:re.split(r'\[', x)[0]
gdp_data.columns = gdp_data.columns.map(column_name_cleaner)
gdp_data.head()| Country/Territory | IMF | World Bank | United Nations | |
|---|---|---|---|---|
| 0 | Monaco | — | 240862 | 240535 |
| 1 | Liechtenstein | — | 187267 | 197268 |
| 2 | Luxembourg | 135321 | 128259 | 125897 |
| 3 | Bermuda | — | 123091 | 117568 |
| 4 | Switzerland | 106098 | 99995 | 93636 |
# set the country column as the index
gdp_data.set_index('Country/Territory', inplace=True)
# convert all other columns into numeric and coerce errors
gdp_data = gdp_data.apply(pd.to_numeric, errors='coerce')
# drop rows with NaN values
gdp_data.dropna(inplace=True)
#find the average GDP per capita
gdp_data['Average GDP per capita'] = gdp_data.mean(axis=1)
gdp_data.head()| IMF | World Bank | United Nations | Average GDP per capita | |
|---|---|---|---|---|
| Country/Territory | ||||
| Luxembourg | 135321.0 | 128259.0 | 125897.0 | 129825.666667 |
| Switzerland | 106098.0 | 99995.0 | 93636.0 | 99909.666667 |
| Ireland | 103500.0 | 103685.0 | 105993.0 | 104392.666667 |
| Norway | 90434.0 | 87962.0 | 106623.0 | 95006.333333 |
| Singapore | 89370.0 | 84734.0 | 78115.0 | 84073.000000 |
12.9.2 Practice exercise 2
Read the spotify dataset from spotify_data.csv that contains information about tracks and artists
12.9.2.1
Find the mean and standard deviation of the track popularity for each genre.
12.9.2.2
Create a new categorical column, energy_lvl, with two levels – ‘Low energy’ and ‘High energy’ – using equal-sized bins based on the track’s energy level. Then, calculate the mean, standard deviation, and 90th percentile of track popularity for each genre and energy level combination
12.9.2.3
Find the mean and standard deviation of track popularity and danceability for each genre and energy level. What insights you can gain from the generated table
12.9.2.4
For each genre and energy level, find the mean and standard deviation of the track popularity, and the minimum and maximum values of loudness.
12.9.2.5
Find the most popular artist from each genre.
12.9.2.6
Filter the first 4 columns of the spotify dataset. Drop duplicate observartions in the resulting dataset using the Pandas DataFrame method drop_duplicates(). Find the top 3 most popular artists for each genre.
12.9.2.7
The spotify dataset has more than 200k observations. It may be expensive to operate with so many observations. Take a random sample of 650 observations to analyze spotify data, such that all genres are equally represented.
12.9.2.8
Find the correlation between danceability and track popularity for each genre-energy level combination.
12.9.2.9
Find the mean of track popularity for each genre-energy lvl combination such that each row corresponds to a genre, and the energy levels correspond to columns.
Hints: using pivot_table()
12.9.2.10
Find the \(90^{th}\) percentile of track popularity for each genre-energy lvl combination such that each row corresponds to a genre, and the energy levels correspond to columns.
Hints: using pivot_table()
12.9.2.11
Find the number of observations in each group, where each groups corresponds to a distinct genre-energy lvl combination
12.9.2.12
Find the percentage of observations in each group of the above table.
12.9.2.13
What percentage of unique tracks are contributed by the top 5 artists of each genre?
Hint: Find the top 5 artists based on artist_popularity for each genre. Count the total number of unique tracks (track_name) contributed by these artists. Divide this number by the total number of unique tracks in the data. The nunique() function will be useful.
